FORGEdb scores are used for predicting which genetic variants are most likely to be a regulatory variant. FORGEdb scores range between 0 and 10, and are calculated using different regulatory DNA datasets, including data for transcription factor (TF) binding and chromatin accessibility.
Specifically, FORGEdb scores are computed from the sum of the presence/absence of the following elements:
A FORGEdb score of 10 (the highest score) is computed from the presence of:
A FORGEdb score of 9 (the second highest score) is computed from the presence of:
A more in-depth explanation of specific scores is available here (with subtitles).
To view a specific example of a SNP with a FORGEdb score of 9, the following figure highlights score details for rs1421085:
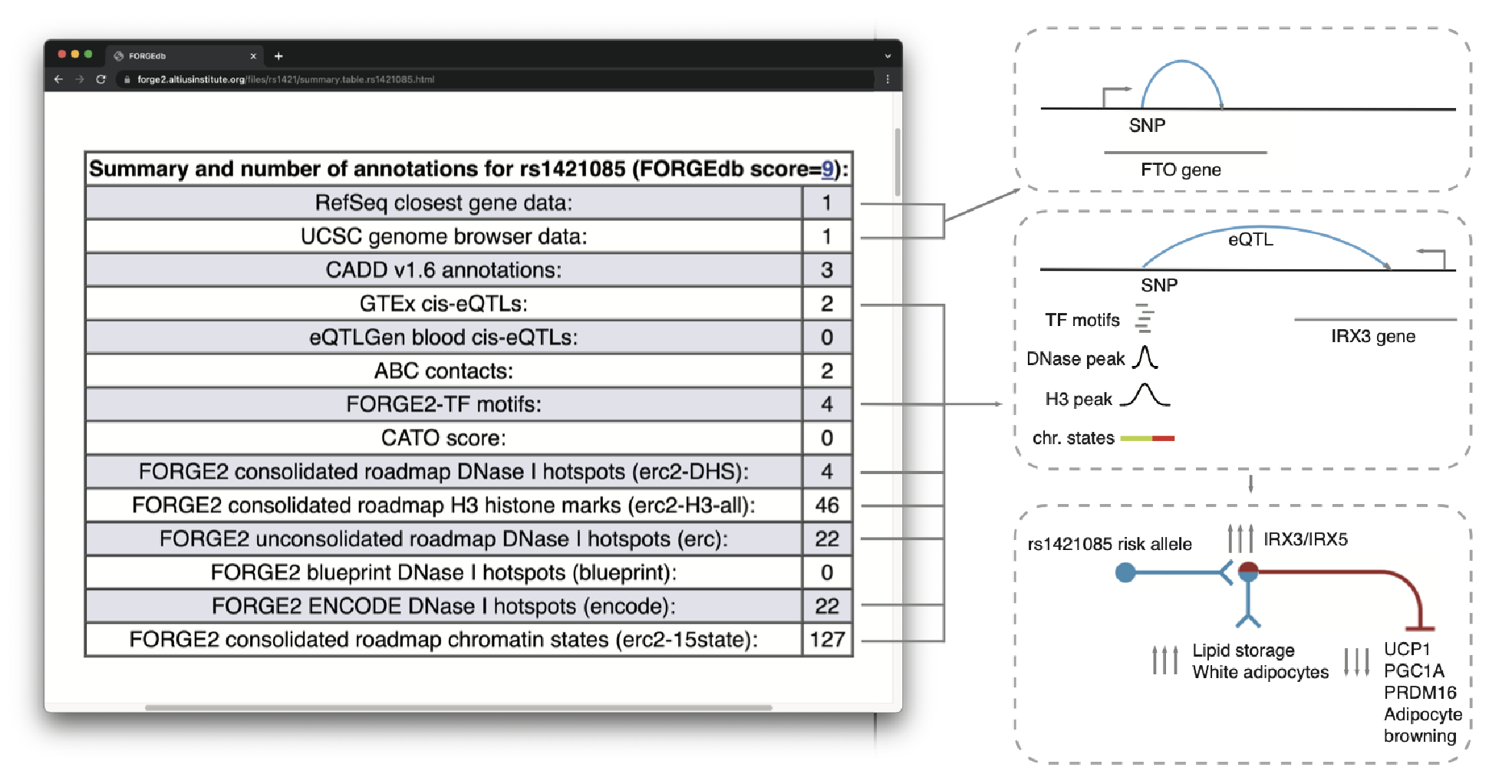
Figure 1: Example FORGEdb results for rs1421085. For this SNP, there is evidence for eQTL associations (with IRX3, shown to be a key target gene by Bell et al. and Smemo et al.), chromatin looping (ABC interactions), overlap with significant TF motifs, DNase I hotspot overlap, as well as overlap with histone mark broadPeaks. The only regulatory dataset that this SNP does not have evidence for is for CATO score (1 point). The resulting FORGEdb score for rs1421085 is therefore 9 = 2 (eQTL) + 2 (ABC) + 1 (TF motif) + 2 (DNase I hotspot) + 2 (histone mark ChIP-seq). Independent experimental analyses by Claussnitzer et al. have demonstrated a regulatory role for this SNP in the regulation of white vs. beige adipocyte proliferation via IRX3/IRX5.
References:
Bell, C. G. et al. Integrated Genetic and Epigenetic Analysis Identifies Haplotype-Specific Methylation in the FTO Type 2 Diabetes and Obesity Susceptibility Locus. PLOS ONE 5, e14040 (2010).
Claussnitzer, M. et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 373, 895–907 (2015).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature, (2014).